Difference between revisions of "Gromacs/Acceleration and parallelization"
From Computational Biophysics and Materials Science Group
(Created page with "Read [http://www.gromacs.org/Documentation/Acceleration_and_parallelization this] first. On Combo, we have compiled several builds of Gromacs. List refers to How to use Com...") |
|||
| Line 26: | Line 26: | ||
Points to note: | Points to note: | ||
* For single machine only, i.e. N<=16 | * For single machine only, i.e. N<=16 | ||
| + | |||
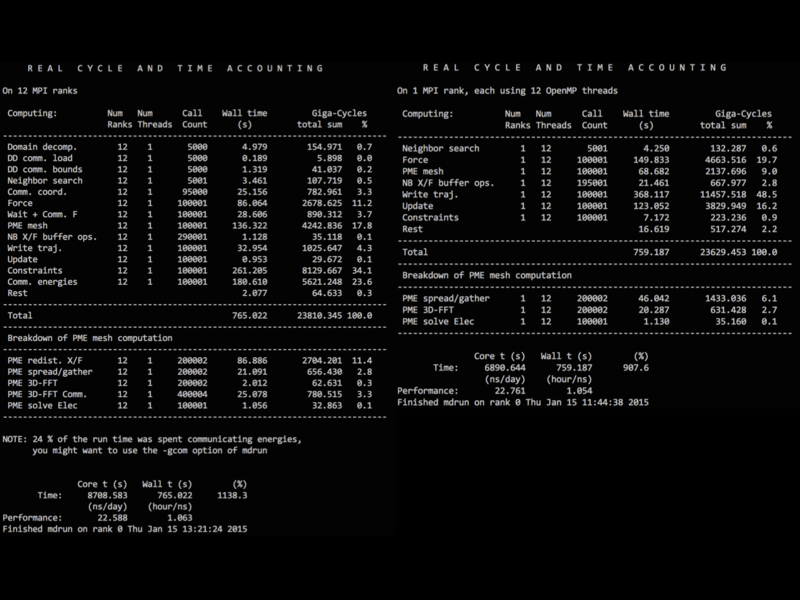
| + | ==Benchmark== | ||
| + | [[File:Compare_mpi.png|800px]] | ||
Revision as of 20:18, 15 January 2015
Read this first.
On Combo, we have compiled several builds of Gromacs. List refers to How to use Combo and compilation options refer to How to compile Gromacs.
We now provide mdrun options for each build:
Contents
[hide]Gromacs 5.0.4 MPI, single, GPU
UNDER TESTS
Gromacs 5.0.4 MPI, double
mpirun -np M mdrun
Points to note: * M is the total number of cores you wish to use * Dunno why M=32 can only utilize one single node, i.e. M=48 utilizes one and half node observed from Ganglia
Gromacs 5.0.4 CUDA, single
mdrun (-nt N) (-pin on)
Points to note: * For single machine only, i.e. N<=16
Gromacs 5.0.4 thread-MPI, double
mdrun (-nt N) (-pin on)
Points to note: * For single machine only, i.e. N<=16